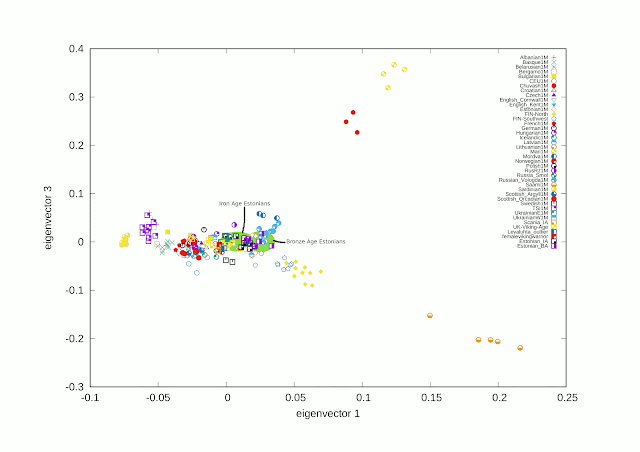
QpAdm was designed to detect admixtures giving also probability and standard error statistics. Two kind of parameters are inputted: admixture candidates and outgroup populations. The result quality depends on both inputted groups. They can be incomplete in many ways. Outgroups should represent some ancestral junctions of tested populations, giving ancestral differences. If there are multiple outgroups representing same ancestral junction they should give a coherent picture. It is not a foregone conclusion in every case because every ancestral branch lives its own history. That is why it is not easy to choose outgroups. Many other problems exist too. If there are in tested groups several ancestrally close populations there will be big standard errors due to common genetic drift of those groups. This means that qpAdm is more suitable for testing very ancient admixtures which are distinctly detectable. Following tests, made using Iron Age populations in the Baltic area, are only directional.
Iron Age Estonians
Estonian_BA / Baltic_IA / Scania_IA
best coefficients: 0.334 0.613 0.053
Jackknife mean: 0.325213405 0.531536401 0.143250193
std. errors: 0.326 0.452 0.490
chisq: 5.922
tail prob: 0.655969
The result imply that the best available result calls for all three ancestral populations, one from Sweden, but sorting of them is difficult due to largely common genetic drift.
Sample 0LS10 (Kunda, Lääne-Viru. EST IA 770–430 BC H13a1a1a N3a3'5)
Estonian_IA / Saami
best coefficients: 0.847 0.153
Jackknife mean: 0.846059225 0.153940775
std. errors: 0.059 0.059
chisq: 4.147
tail prob: 0.901439
The standard error is much lower due to lesser common genetic drift. In this case the total error is probably generated by the Finnish admixture in present-day Saami samples. The eastern admixture of 0LS10 is very purely eastern and barely shows any Finnish ancestry. Would I say that it is question about proto-Saami ancestry...
Finnish genetics / older posts
lauantai 28. joulukuuta 2019
keskiviikko 11. joulukuuta 2019
New "Viking sword find" in North Estonia turned out to be of local origin?
I read about this find a month ago and my first thought was that yes, has to be Finnish. Why? Just because Finland is the nearest place where we have seen plenty of these swords and connections between Finland and North Estonia were still tight during that time. Now also Estonian research deny the Scandinavian origin. One reason for this new idea was the find of a brooch with typical Finnish-North Estonian shape. Not much, but obviously something more is to be published sooner or later. Especially so called crawfish brooches were distinctive in Finland, but I have to wait more detailed pictures before saying more because Estonian vocabulary can differ from what I have seen. The new article states:
"Crossbow-shaped brooches were usually worn by warriors from southwestern Finland and northwestern Estonia on the passage that intersects with the main thoroughfare of the Eastern Route," Kiudsoo said."
I'll wrote more after more details are revealed, including pictures of those finds.
https://news.err.ee/1012100/burial-site-ties-major-viking-swords-find-to-warriors-from-ravala
"Crossbow-shaped brooches were usually worn by warriors from southwestern Finland and northwestern Estonia on the passage that intersects with the main thoroughfare of the Eastern Route," Kiudsoo said."
I'll wrote more after more details are revealed, including pictures of those finds.
https://news.err.ee/1012100/burial-site-ties-major-viking-swords-find-to-warriors-from-ravala
lauantai 7. joulukuuta 2019
Increasing western influence in Iron Age Estonia
Despite of premature rumors of eastern influence in Iron Age Estonia the change in the beginning of the Iron Age came from the west. Saag et al. 2019 seemingly gives contradictory information, because given samples tell another story. I grouped those samples into two groups depending on sample timings.
Estonian BA
Estonian_BA V9_2 1060–850 BC R1a1’2
Estonian_BA V14_2 1280–1050 BC R1a1’2
Estonian_BA V16_1 730–390 BC R1a1’2
Estonian_BA X11_1 1030–890 BC R1a
Estonian_BA X14_1 780–430 BC R1a1c
Estonian_BA X20_1 900–800 BC R1a
Estonian IA
Estonian_IA V11_1 390–200 BC -
Estonian_IA V12_1 360–40 BC N3a3a
Estonian_IA VII3_1 380–180 BC ?
Estonian_IA VIII5_2 75–300 AD R1a
Estonian_IA VIII7_1 75–200 AD -
Estonian_IA VIII8_1 75–200 AD R1a1c
Estonian_IA VIII9_1 75–200 AD -
I made the following PCA to fulfill needs of those who are interested in Siberian admixtures, want to see possible Siberian connections in context of Baltic Finnic and Volga Finnic languages in terms of genes if it is even possible. PCA plots are always made for certain purposes and every individual plot tells certain true story, but never the full story.
Notice also that so called Levanluhta outlier found from Iron Age Ostrobothnia fits well with the Viking group and Vikings don't match well with present-day Scandinavian and British people.
Update 9.12.2019 21:00
Now added a PCA plot figuring ancient Estonian N1c1 samples.
0LS10 Kunda Lääne-Viru, EST IA 770–430 BC M XY H13a1a1a N3a3 0 5
V12 Kurevere, Saare, EST IA 360–40 BC M XY I1a1c N3a3a
VII4 Vohma lääne-Viru, EST IA 760–400 BC M XY T1a1b N3a3a
IIa Karja, Saare, EST MA 1230–1300 AD M XY H3h1 N3a3a
IIf Otepää, Valga, EST MA 1360–1390 AD M XY T2b N3a3a
IIg Pada, Lääne-Viru, EST MA 1210–1230/1240 AD M XY U4a2b N3a3a
0LS10 drifts towards Siberian admixed groups and IIa drifts towards Finns and Saamis. Both deviants are however too weak to be considered as real migrants and their locations probably represent only admixture. All N1c1 samples are more comparable to those more western Iron Age Estonians than to more Baltic-like Bronze Age Estonians.
Estonian BA
Estonian_BA V9_2 1060–850 BC R1a1’2
Estonian_BA V14_2 1280–1050 BC R1a1’2
Estonian_BA V16_1 730–390 BC R1a1’2
Estonian_BA X11_1 1030–890 BC R1a
Estonian_BA X14_1 780–430 BC R1a1c
Estonian_BA X20_1 900–800 BC R1a
Estonian IA
Estonian_IA V11_1 390–200 BC -
Estonian_IA V12_1 360–40 BC N3a3a
Estonian_IA VII3_1 380–180 BC ?
Estonian_IA VIII5_2 75–300 AD R1a
Estonian_IA VIII7_1 75–200 AD -
Estonian_IA VIII8_1 75–200 AD R1a1c
Estonian_IA VIII9_1 75–200 AD -
Update 9.12.2019 21:00
Now added a PCA plot figuring ancient Estonian N1c1 samples.
0LS10 Kunda Lääne-Viru, EST IA 770–430 BC M XY H13a1a1a N3a3 0 5
V12 Kurevere, Saare, EST IA 360–40 BC M XY I1a1c N3a3a
VII4 Vohma lääne-Viru, EST IA 760–400 BC M XY T1a1b N3a3a
IIa Karja, Saare, EST MA 1230–1300 AD M XY H3h1 N3a3a
IIf Otepää, Valga, EST MA 1360–1390 AD M XY T2b N3a3a
IIg Pada, Lääne-Viru, EST MA 1210–1230/1240 AD M XY U4a2b N3a3a
0LS10 drifts towards Siberian admixed groups and IIa drifts towards Finns and Saamis. Both deviants are however too weak to be considered as real migrants and their locations probably represent only admixture. All N1c1 samples are more comparable to those more western Iron Age Estonians than to more Baltic-like Bronze Age Estonians.
sunnuntai 17. marraskuuta 2019
Mitochondrial lineages in Iron Age Fennoscandinavia, comments
The distribution between lines U and H in Finland has been a point of discussion now. Many thinkers have underlined that in a big view H is connected to Neolithic farmers and U to Mesolithic hunter gatherers. Although it is interesting to combine different point of views, sometimes people forget the context and tend to overreact, as in this case has happened too. Overall the transition to farming happened in Southern Europe around 7000-5000 BCE and in Northern Europe 3000-2000 BCE. In light of this history all settlements in Finland are extremely young and in Southern Finland differences between female and male haplogroups manifest more migration routes from different compass points and different farmer populations, rather than introduction of Finnish agriculture. Although I don't believe that this fact is totally missed, wrong highlighting can't be any good. Populations and migrations varied depending on time, during the latest 2000 years especially in the East Europe. In my opinion in this point researchers have lost the substance, which would hold a question "where did they come and why so much later than in most other places in Europe". The answer is to be discovered by comparing female and male haplogroups and types in Finland and in possible contact areas.
lauantai 16. marraskuuta 2019
Mitochondrial lineages in Iron Age Fennoscandinavia
A new study has been published, Human mitochondrial DNA lineages in Iron-Age Fennoscandia suggest incipient admixture and eastern introduction of farming-related maternal ancestry . It includes several new samples from Iron Age Finland and also historical Finnish samples for a comparison purpose. Some of these mitochondrial samples are far too rare to reveal statistically anything, some are too common. Best we can do is to try to find results implying something regional and I try to find them and compare them to present-day samples gathered from FamilyTreeDna's projects. Unfortunately those projects are biased to West Europe due to the testing activity, yet we can see distribution patterns inside Fennoscandinavia, because Finland is one of the best tested country. I am not going to make conclusions, because I don't see any sense to try to connect only 1000 years old Finnish samples to any European wide prehistoric events, I just show results. When I have more time I will gather more results, if it sounds reasonable in a Fennoscandinavian regional context. You can do your comparisons and send here as comments. Image cropping follows the distribution, nothing exists outside images.
Levänluhta2 JK1963 U5b1b1a1
Levänluhta5 JK1966 U5b1b1a1a
Levänluhta8 JK1969 U5b1b1a1b
Levänluhta11 JK2066 U5b2a5(a)
Hollola8 TU662 U4b1b1
Luistari1 TU464 U4b1b1a
Hollola14 TU670 U4b1b1b
Hollola6 TU660 U4d1a1a
Hollola10 TU664 U4a2
Hollola15 TU672 V7a
Tuukkala7 TU643 V7a1
Tuukkala10 TU646 H1a7
Hiitola11 TU676 H1a7
Hiitola8 TU570 H1b2
Hiitola1 TU504 H1a8a
Tuukkala5 TU633 H1a8a
Hiitola4 TU564 H2a2a1b
Tuukkala6 TU641 H11a1
Levänluhta2 JK1963 U5b1b1a1
Levänluhta5 JK1966 U5b1b1a1a
Levänluhta8 JK1969 U5b1b1a1b
Levänluhta11 JK2066 U5b2a5(a)
Hollola8 TU662 U4b1b1
Luistari1 TU464 U4b1b1a
Hollola14 TU670 U4b1b1b
Hollola6 TU660 U4d1a1a
Hollola10 TU664 U4a2
Hollola15 TU672 V7a
Tuukkala7 TU643 V7a1
Tuukkala10 TU646 H1a7
Hiitola11 TU676 H1a7
Hiitola8 TU570 H1b2
Hiitola1 TU504 H1a8a
Tuukkala5 TU633 H1a8a
Hiitola4 TU564 H2a2a1b
Tuukkala6 TU641 H11a1
keskiviikko 6. marraskuuta 2019
Challenge youself, things aren't always piece of cake
Behind most studies and also behind yDna haplotree nomenclatures is a lot of brain work. Just reading and picking scientific vocabulary doesn't make you an expert, you have to experience things. See differences and similarities, question your own thoughts more than thoughts of other people and question enough to reach consistent structures. These comments are not addressed to anyone particularly, but something I have seen has to be commented.
I promised to go on with my yDna tests and add also "negative calls", i.e. ancestral SNPs. I have retested Stora Förvar 11, a sample from Gotland dated 7300-6800 BC and detected by me and another blogger as I1 . Before new results some background. If we look at the ISOGG 2019 nomenclature we see a significant difference between branches I1 and I2. There are 205 found positive calls under the classification I1, but only 22 under I2. I counted now only calls under titles I1 and I2, not any downstream mutations. This means that the time span in case of the title I1 is bigger and it also means that there is a lot of unknown history behind the I1 story. So when we say "it is only a title I1" we actually should say "it is I1 and many still unknown downstream mutations", almost 10 times more time and unknown downstream mutations than in case of I2. We don't know much about the prehistory of I1 until we have dated information of its "positive calls".
There were "positive calls" / derived values for Stora Forvar 11:
and there are "negative calls" / ancestral values:
We can say that Stora Förvar 11 belongs to the I1 branch and if we could infer the age he could be located around to the middle of the time span of the title level of I1 (11 derived and 10 ancestral SNPs). All ISOGG locations above I1 in I branch were derived and all below I1 were ancestral.
I promised to go on with my yDna tests and add also "negative calls", i.e. ancestral SNPs. I have retested Stora Förvar 11, a sample from Gotland dated 7300-6800 BC and detected by me and another blogger as I1 . Before new results some background. If we look at the ISOGG 2019 nomenclature we see a significant difference between branches I1 and I2. There are 205 found positive calls under the classification I1, but only 22 under I2. I counted now only calls under titles I1 and I2, not any downstream mutations. This means that the time span in case of the title I1 is bigger and it also means that there is a lot of unknown history behind the I1 story. So when we say "it is only a title I1" we actually should say "it is I1 and many still unknown downstream mutations", almost 10 times more time and unknown downstream mutations than in case of I2. We don't know much about the prehistory of I1 until we have dated information of its "positive calls".
There were "positive calls" / derived values for Stora Forvar 11:
| FGC2433 | I1 |
| CTS1748 | I1 |
| Z2749 | I1 |
| CTS6221^ | I1 |
| Z2802^ | I1 |
| CTS6629 | I1 |
| Z2805 | I1 |
| YSC0000301 | I1 |
| Z2882 | I1 |
| Z2726 | I1 |
| Z2731 | I1 |
and there are "negative calls" / ancestral values:
| CTS3268 | I1 |
| Z2762 | I1 |
| L121 | I1 |
| S62 | I1 |
| CTS9258 | I1 |
| Z2825 | I1 |
| L509 | I1 |
| Z2864 | I1 |
| FGC2425 | I1 |
| Z2714 | I1 |
We can say that Stora Förvar 11 belongs to the I1 branch and if we could infer the age he could be located around to the middle of the time span of the title level of I1 (11 derived and 10 ancestral SNPs). All ISOGG locations above I1 in I branch were derived and all below I1 were ancestral.
sunnuntai 3. marraskuuta 2019
Thoughts about the origin of Finnish and Estonian languages
This is probably not a popular writing, but I am not a politician and I don't need to please anyone. Here is the issue: why the yDna distribution among ancestors of the earliest Finnic people doesn't correspond to their imagined yDna? Linguists are cosily unanimous that the home of Baltic Finnic languages was in Southern Estonia around 3000 years ago. One but not the only reason for this has been the presence of Baltic loan words in Baltic Finnic languages, including Finnish. Numerous studies endorse this idea. Baltic loan words wouldn't have been possible without straight connection with Baltic speakers. But then, why this theory is not supported by genetics? It is a common idea that the haplogroup N1c1 brought Finnic languages from the east to Estonia. However, only a small minority of Late Bronze Age and Iron Age men in Estonia carried N1c1 and even less of them carried those clades nowadays especially connected to Baltic Finnic speakers. Below statistics from Saag et al. 2019. 21 samples of all 28 ancient Estonians carried R1a and only 6 belonged to N1c1, one was J2b2. Furthermore only one of those six belonged to a "Finnish haplotype" and other five most likely belonged to "Baltic haplotypes".
Tilaa:
Kommentit (Atom)
Some ancestral changes in Iron Age Estonia
QpAdm was designed to detect admixtures giving also probability and standard error statistics. Two kind of parameters are inputted: admixtu...
-
This is probably not a popular writing, but I am not a politician and I don't need to please anyone. Here is the issue: why the yDna d...
-
Testing ancient "steppe" samples on PCA together with modern ones revealed unexpected issues. Studies have included different set...
-
A new study has been published, Human mitochondrial DNA lineages in Iron-Age Fennoscandia suggest incipient admixture and eastern introduct...